Bio
Hello! I have earned a dual B.S. in Physics and Pharmaceutics at Zhejiang University in 2022, advised by Prof. Tingjun Hou, focusing on Computer-aided Drug Design After that I am involved in a Master program at Zhejiang University and got an internship in Carbon Silicon AI, a startup company, continuing my research in AI-aided Drug Design. In the future, I will go to UW Allen School for my Ph.D. study, advised by Prof. David Baker for ligand drug design. My CV could be found here.
My research interests are computer-aided drug design. Specifically, I am currently working on (1) structure-based drug design methodology (molecular generation and virtual screening approaches) (2) generative modeling (3) molecular dynamics. Basically, I’m also an AI4Science researcher.
News
Publications [Google Scholar]
* indicates equal contribution.Molecular Generation
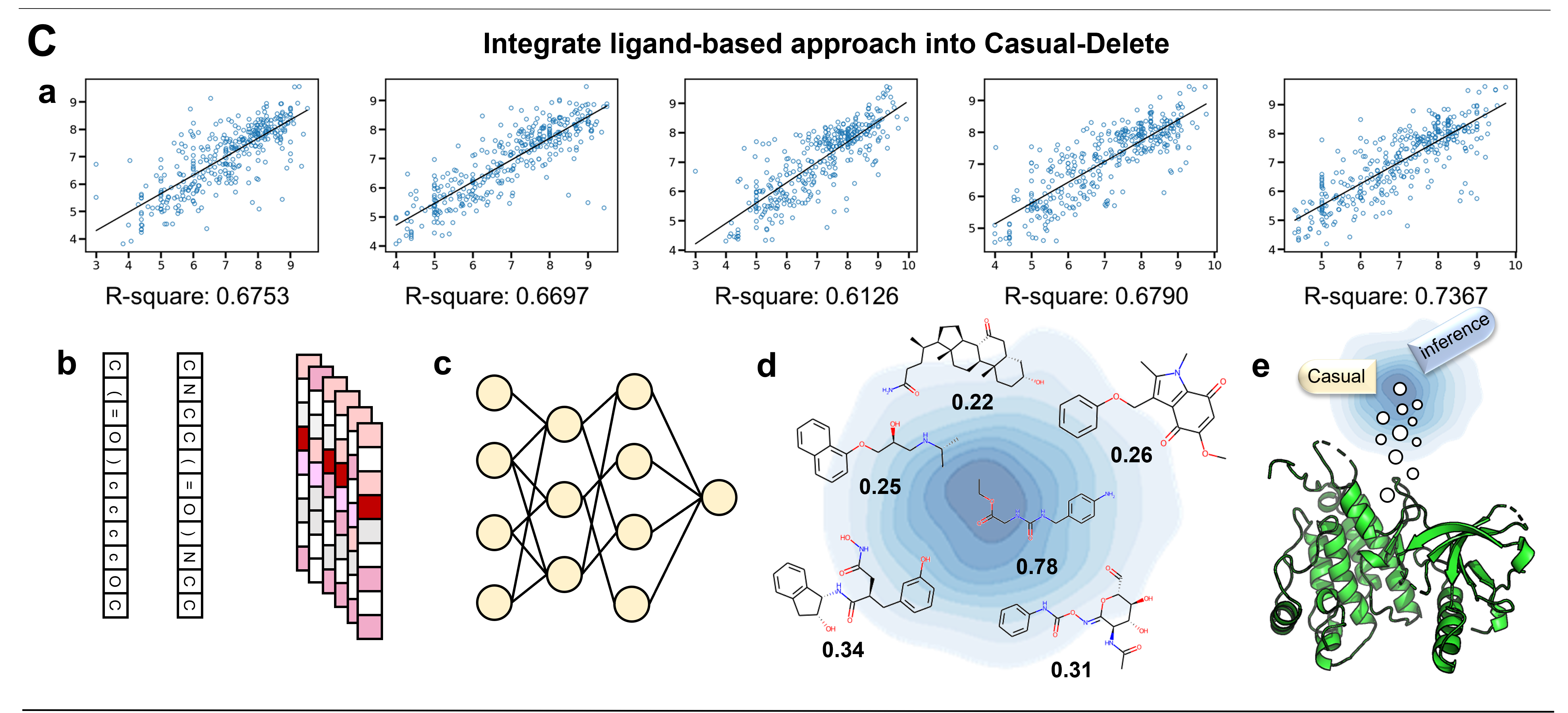
Structure-based Molecular Generation: A New Paradigm Enables Efficient Design of Potent CLIP1-LTK Inhibitors
Peichen Pan*, ShiCheng Chen*, Odin Zhang*, ..., Tingjun Hou
Cell, Submitted
We extend the Delete model to Casual-Delete model, scussfully design a potent inhibitors targeting to LTK
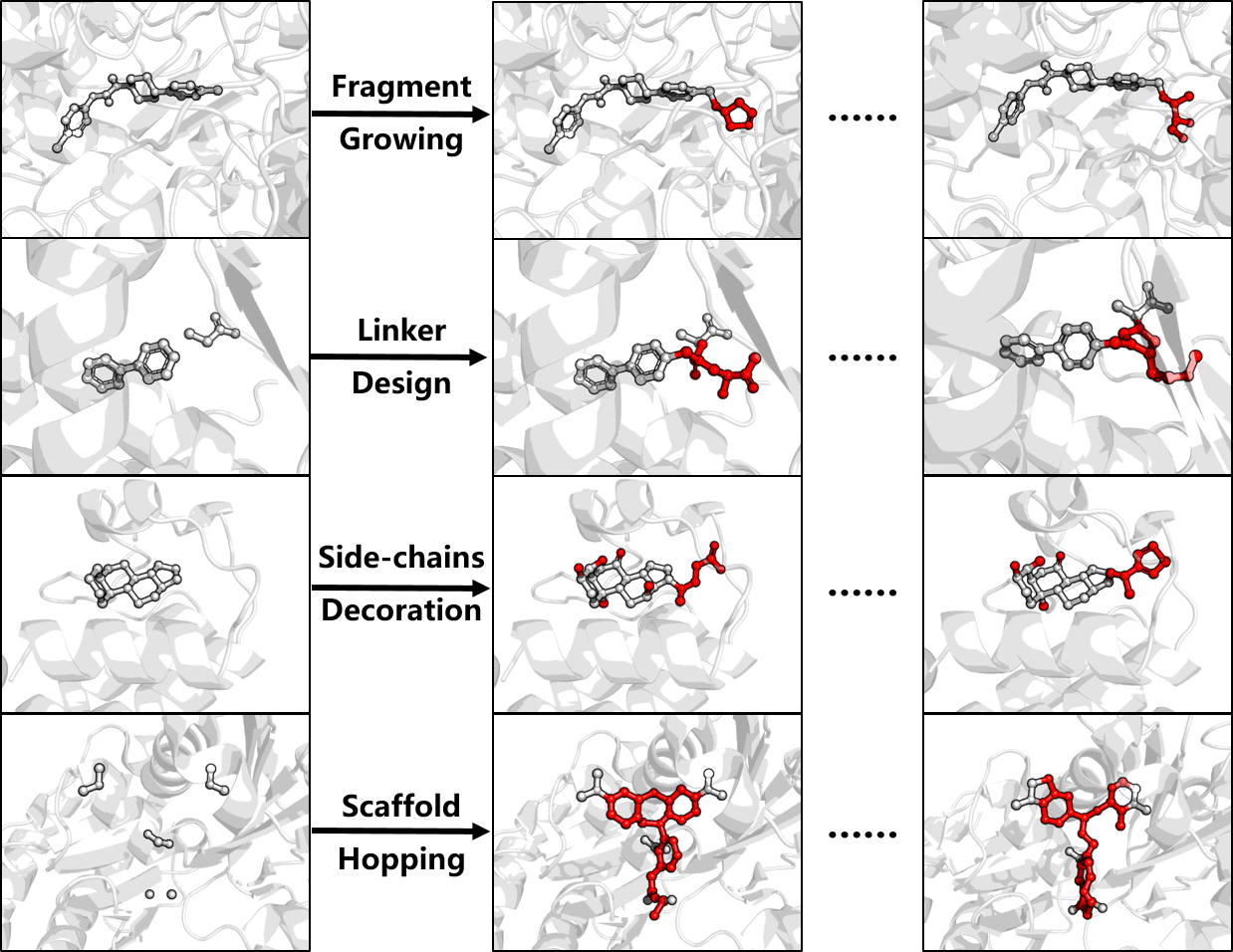
Delete: Deep Lead Optimization Enveloped in Protein Pocket through unified Deleting Strategies and a Structure-aware Network
Odin Zhang*, Huifeng Zhao*, Xujun Zhang*, ..., Chang-yu Hsieh, Tingjun Hou.
Nat. Commun. Second-round Peer Review
When you face some problems in drug discovery, just delete!
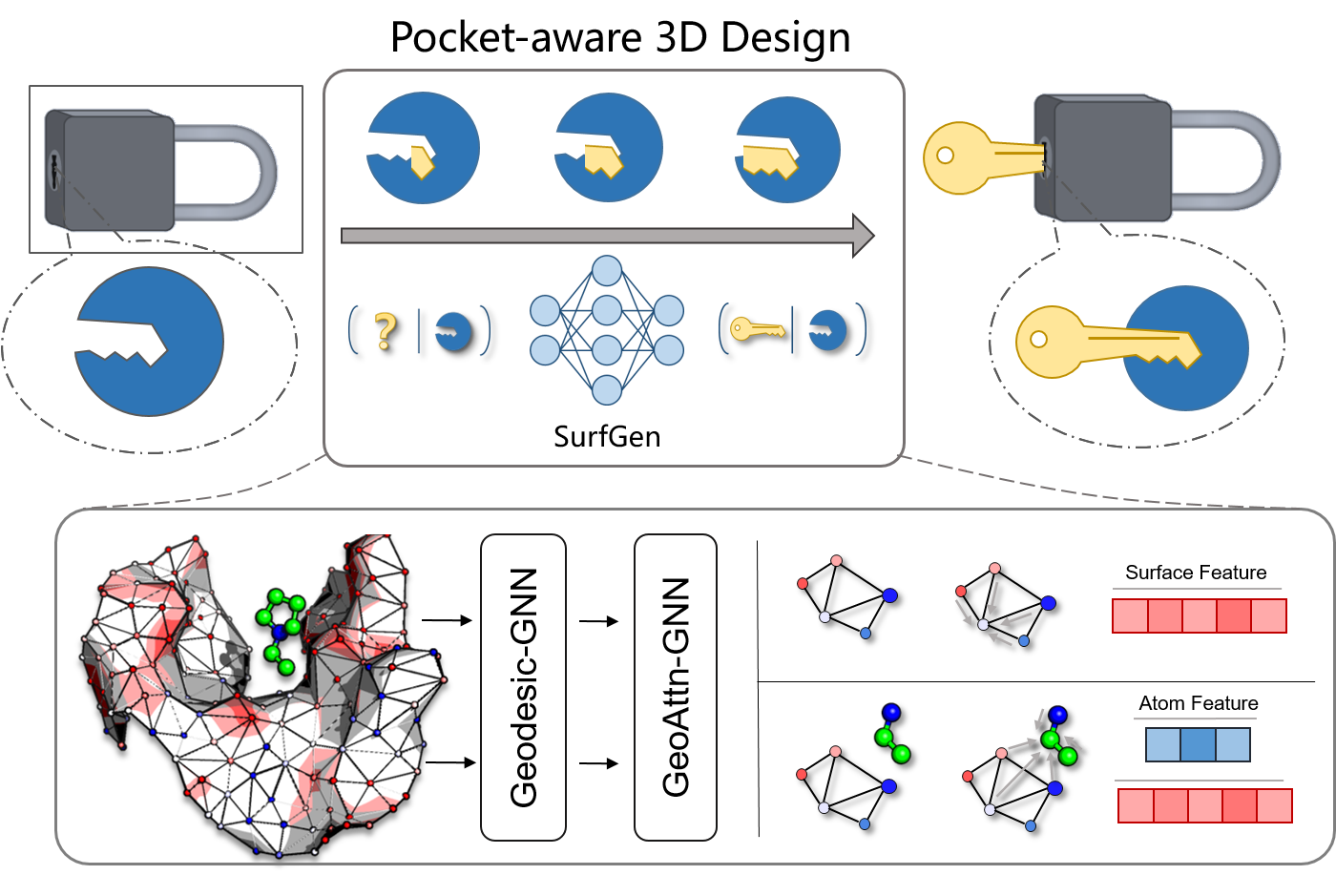
SurfGen: Learning on Topological Surface and Geometric Structure for 3D Molecular Generation
Odin Zhang*, Tianyue Wang*, Gaoqi Weng, ..., Chang-yu Hsieh, Tingjun Hou.
Nat. Compt. Sci, 3, 849–859 (2023)
Inspired from key-and-lock model, we build a 3D molecular design model directly on topological protein surface through Geodesic-GNN and Geoatton-GNN, which satisfy the physical equivariance.
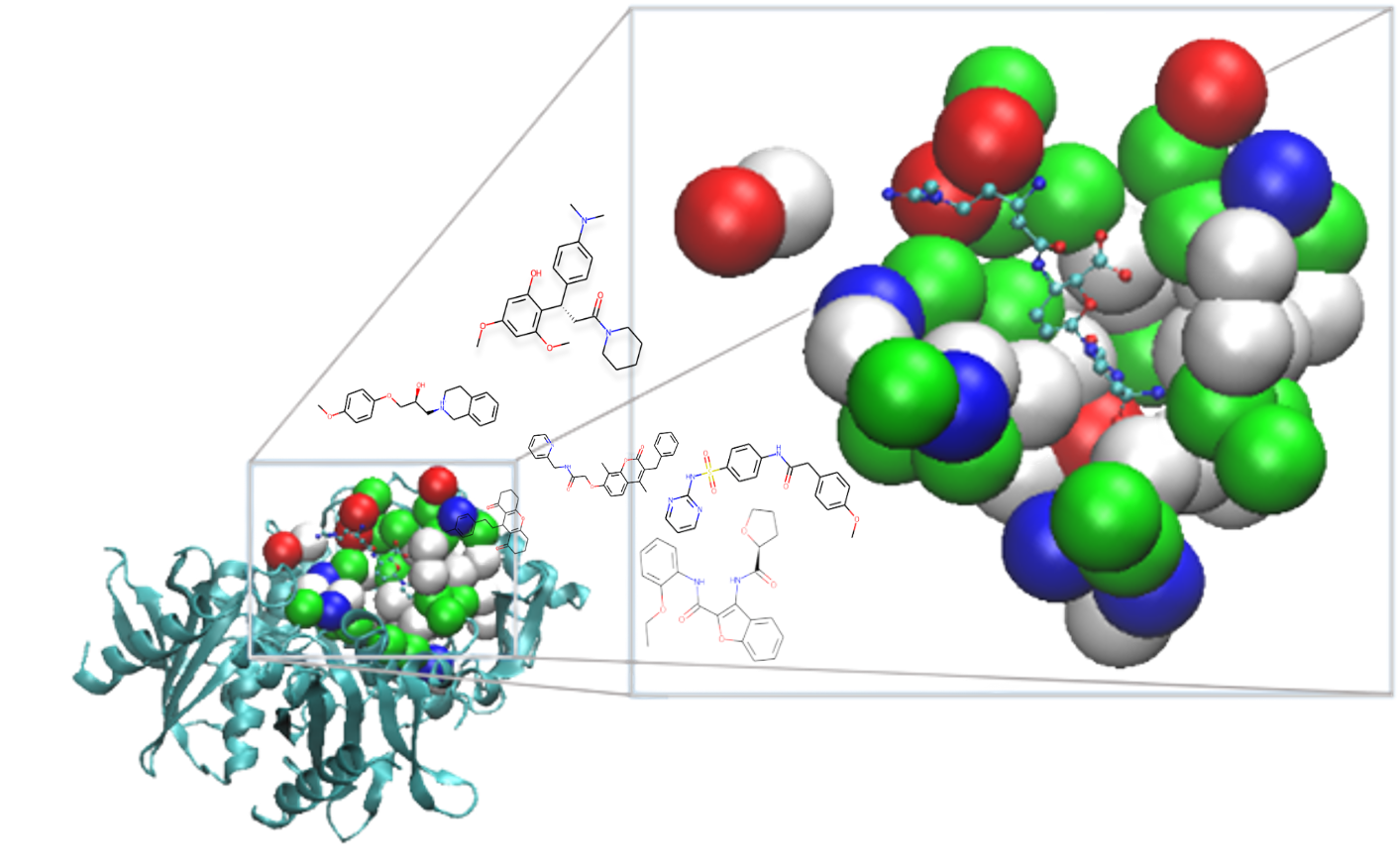
ResGen: Learning the Interaction Patterns through Multiscale Modeling and SE(3)-Framework for 3D-Pocket Molecular Generation
Odin Zhang, Jintu Zhang, Xujun Zhang, ..., Chang-yu Hsieh, Tingjun Hou.
Nat. Mach. Intell., 5, 1020–1030 (2023)
We construct a novel 3D Pocket-aware molecular generation model via multi-scale modeling, achieving SOTA performance on CrossDock dataset and our curated real-world dataset.
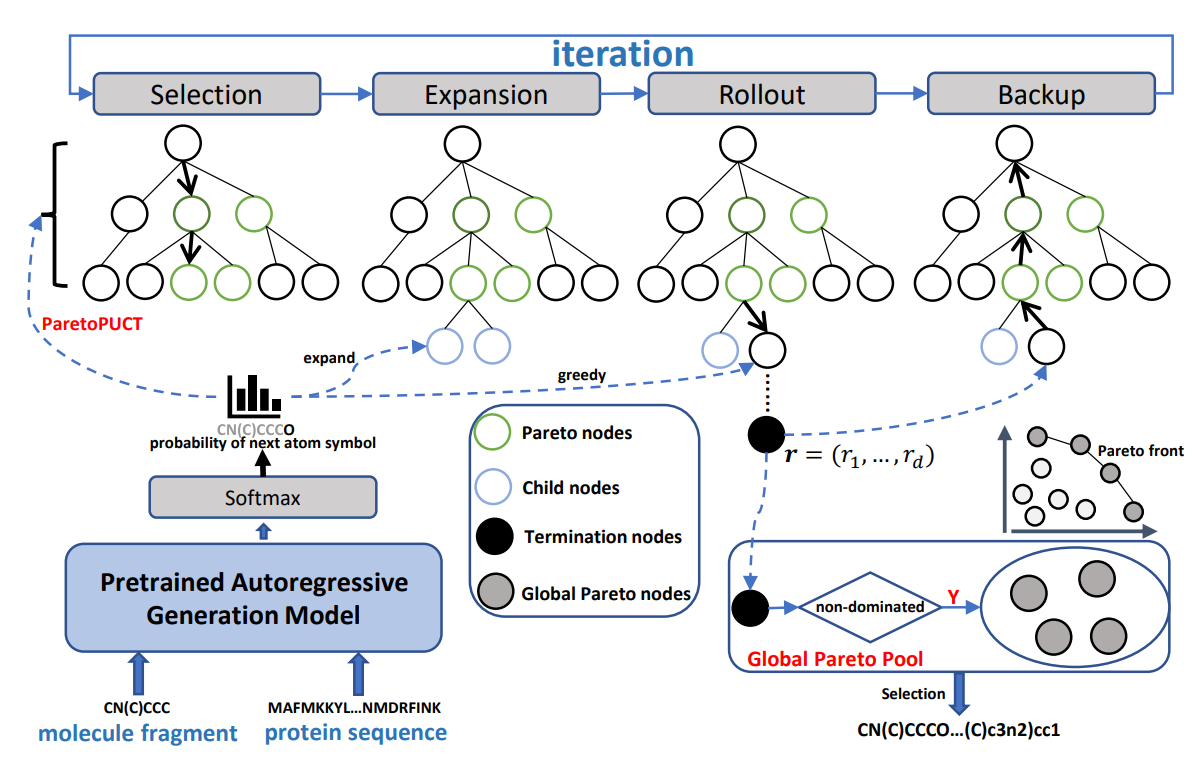
Multi-Objective Structure-Based Molecule Generation with Pareto MCTS
Yaodong Yang*, Guangyong Chen, Jinpeng Li, Odin Zhang*, ..., Pheng-Ann Heng
AAAI
A MCTS-based method for molecular generation
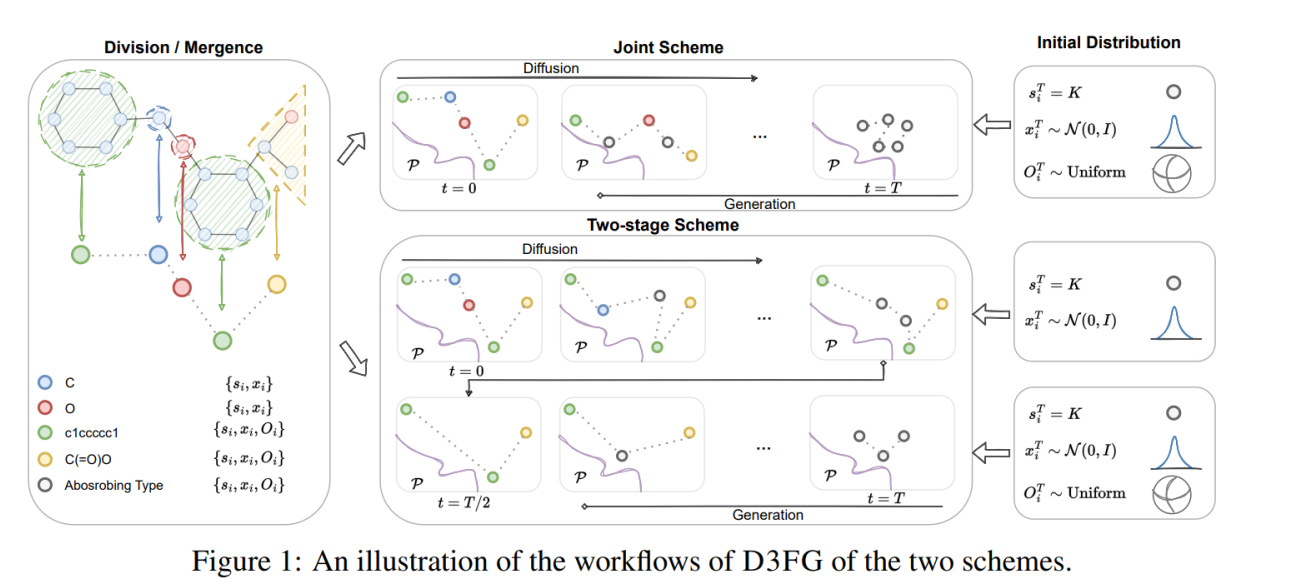
Functional-Group-Based Diffusion for Pocket-Specific Molecule Generation and Elaboration
Haitao Lin, Yufei Huang, Odin Zhang, ..., Stan Z. Li.
NIPS2023
We use diffusion model to grow fragments and link fragments for lead optimization
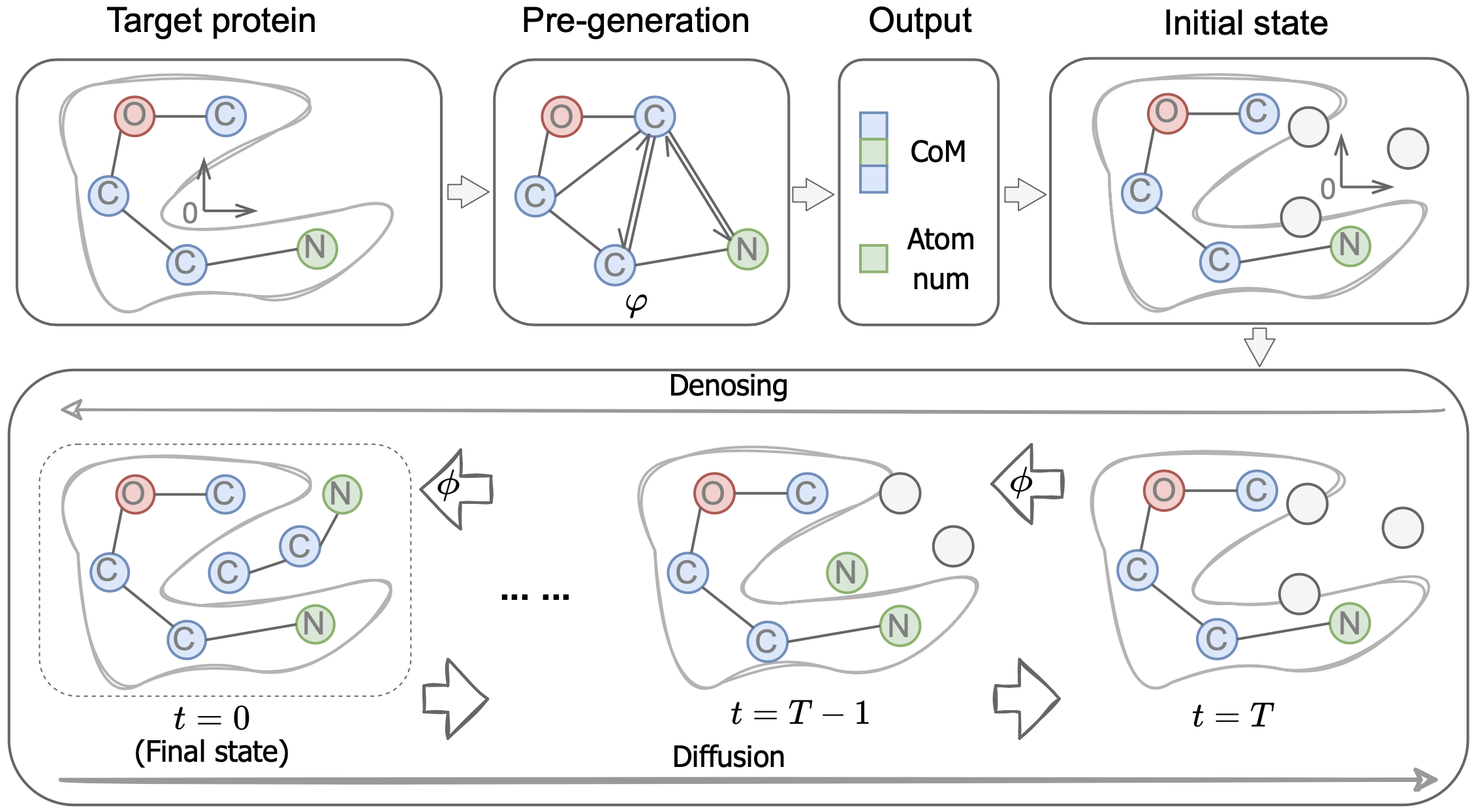
DiffBP: Generative Diffusion of 3D Molecules for Target Protein Binding
Haitao Lin*, Yufei Huang*, Liu Meng, Odin Zhang, Xuanjing Li, Shuiwang Ji, Tingjun Hou, Stan Z. Li
We propose a one-shot approach to capture global interatomic forces for growing molecules directly within protein pockets
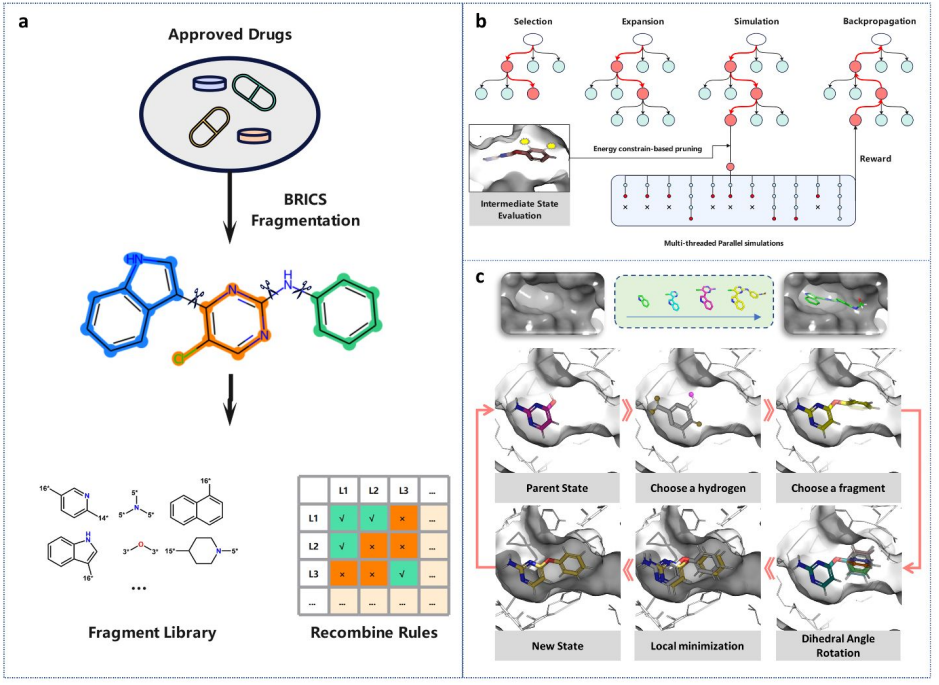
A Flexible Data-Free Framework for Structure-Based De Novo Drug Design with Reinforcement Learning
Hongyan Du*, Dejun Jiang*, Odin Zhang, ..., TingjunHou.
Chemical Science
A MCTS-based approach enables de novo design according to the chemicts' requirement
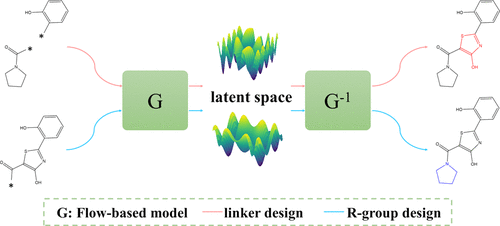
FFLOM: a flow-based autoregressive model for fragment-to-lead optimization. Journal of Medicinal Chemistry
Jieyu Jin, Dong Wang, Odin Zhang, ..., Yu Kang, Tingjun Hou
Journal of Medicinal Chemistry 14(8), 2054-2069
We develop a flow-based model for fragment elaboration and linker design, achieving the SOTA performance regarding 2D metrics
Other AIDD approaches
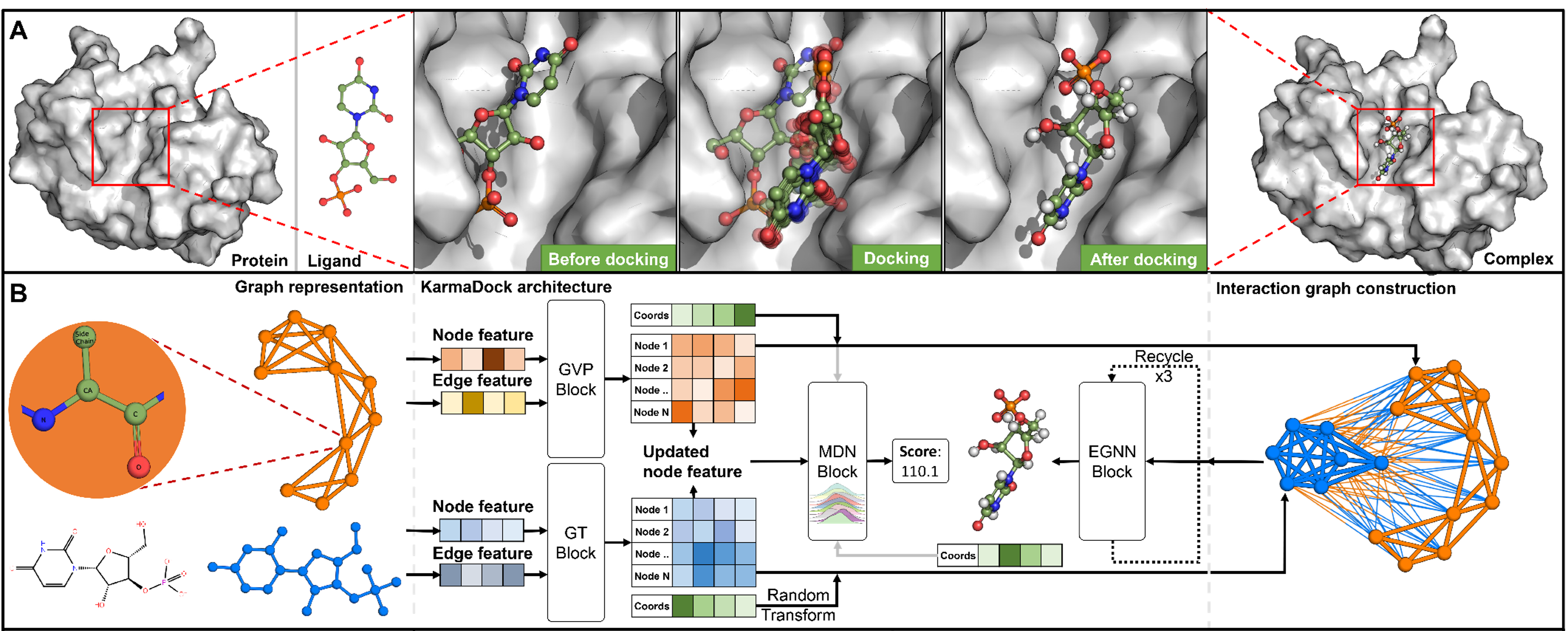
KarmaDock: a deep learning paradigm for ultra-large library docking with fast speed and high accuracy
Xujun Zhang*, Odin Zhang*, ..., Chang-yu Hsieh, Tingjun Hou.
Nat. Compt. Sci.,3, 739–740 (2023).
A Deep Learning-based docking method, from binding pose to virtual screening
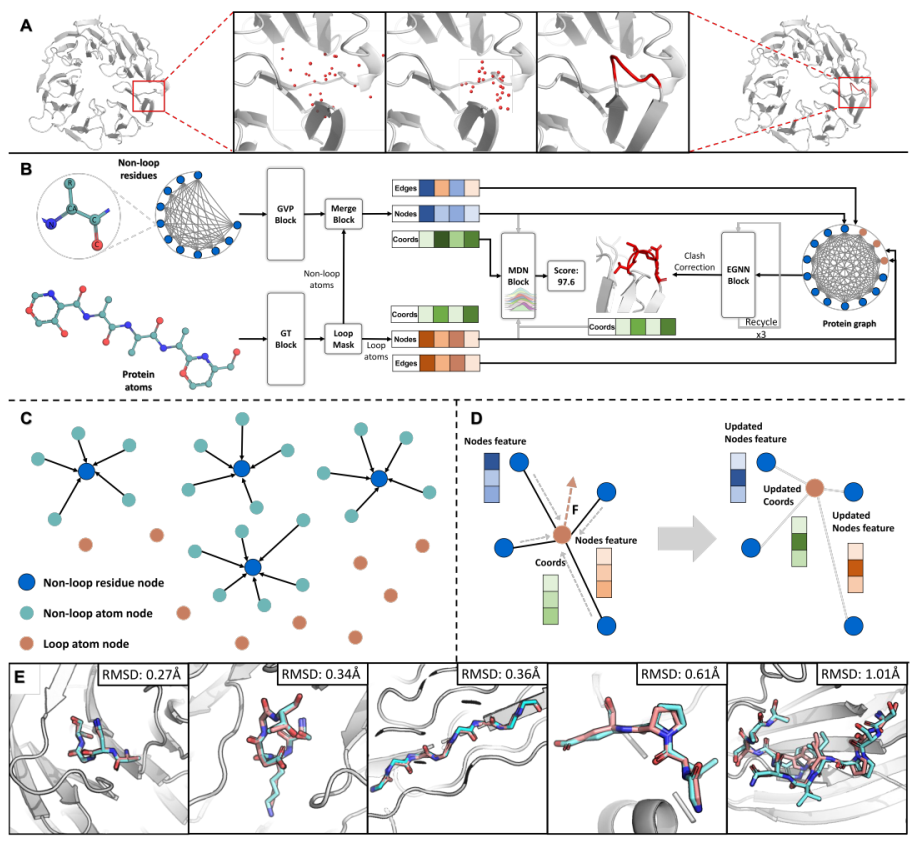
Highly accurate and efficient deep learning paradigm for full-atom protein loop modeling with KarmaLoop
Tianyue Wang*, Xujun Zhang*, Odin Zhang*, ..., Chang-yu Hsieh, Tingjun Hou.
Nat. Mach. Intell., Major Revision
A SOTA method for the geometry prediction of the loop domain in proteins
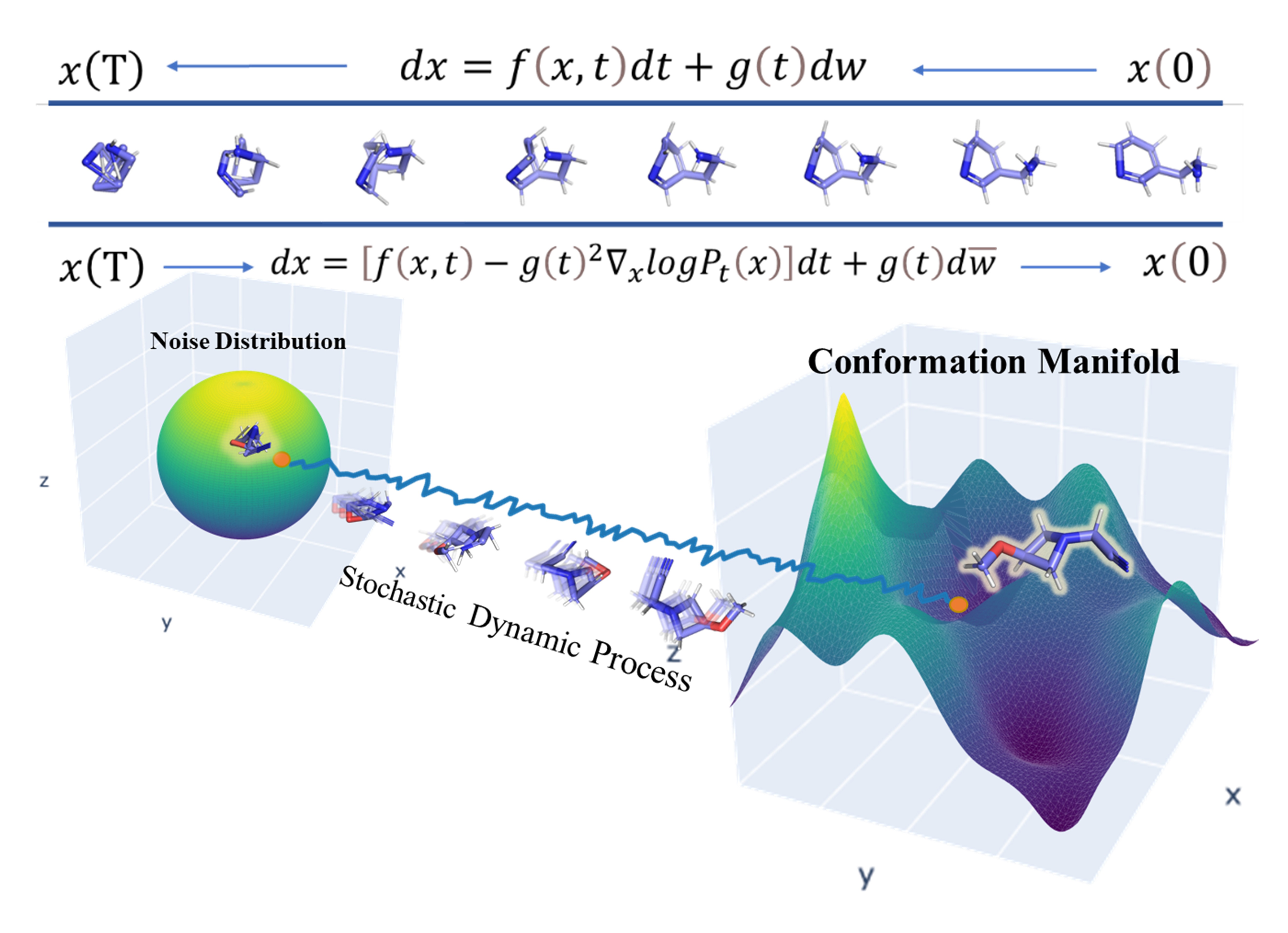
SDEGen: Learning to Evolve Molecular Conformations from Thermodynamic Noise for Conformation Generation
Odin Zhang*, ShengMing Li*, Jintu Zhang, .., Yu Kang, Tingjun Hou.
Chem. Sci., 14, 1557-1568
We construct a novel conformation generation model via the stochastic differential equation-based generative model, which is a kind of diffusion process.
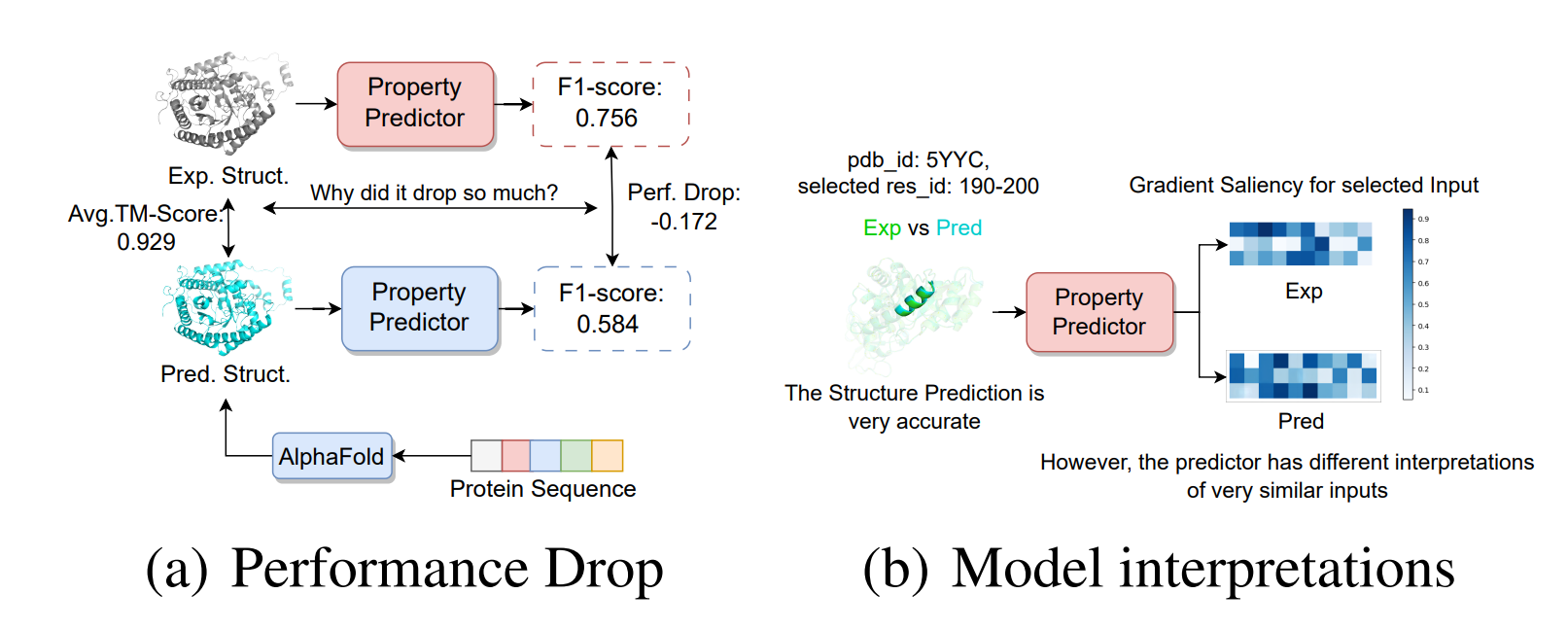
Protein 3D Graph Structure Learning for Robust Structure-based Protein Property Prediction
Yufei Huang*, ..., Odin Zhang, ..., Stan. Z. Li.
AAAI 2024
In this paper, we first investigate the rea- son behind the decrease in prediction accuracy when utilizing predicted structures, attributing it to the structural distribution bias from the perspective of structure representation learning

Infinite Physical Monkey: Do Deep Learning Methods Really Perform Better in Conformation Generation?
Odin Zhang, Jintu Zhang, Huifeng Zhao, Dejun Jiang, Yafeng Deng.
Bio Arxiv
We present our opinions on conformation generation, discussing another two papers on arxiv platform
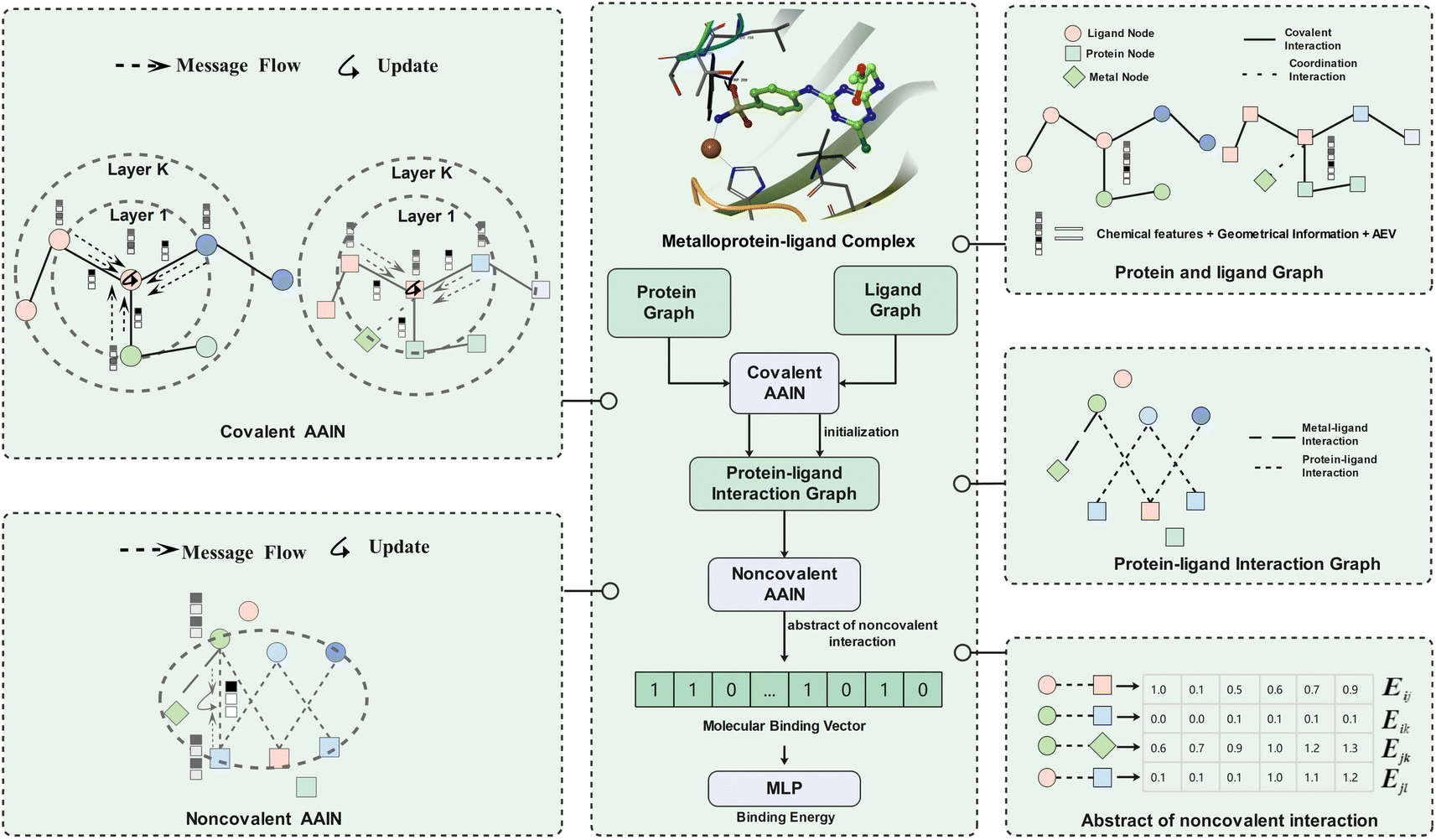
MetalProGNet: A Structure-based Deep Graph Model for Metalloprotein–ligand Interaction Predictions
Dejun Jiang, Zhaofeng Ye, Chang-yu Hsieh, Odin Zhang, ..., Jian Wu, Tingjun Hou
Chem. Sci. 14(8), 2054-2069
We develop a novel and accurate scoring function for metalloproteins, the main methodology is bi-partite graph and GNN
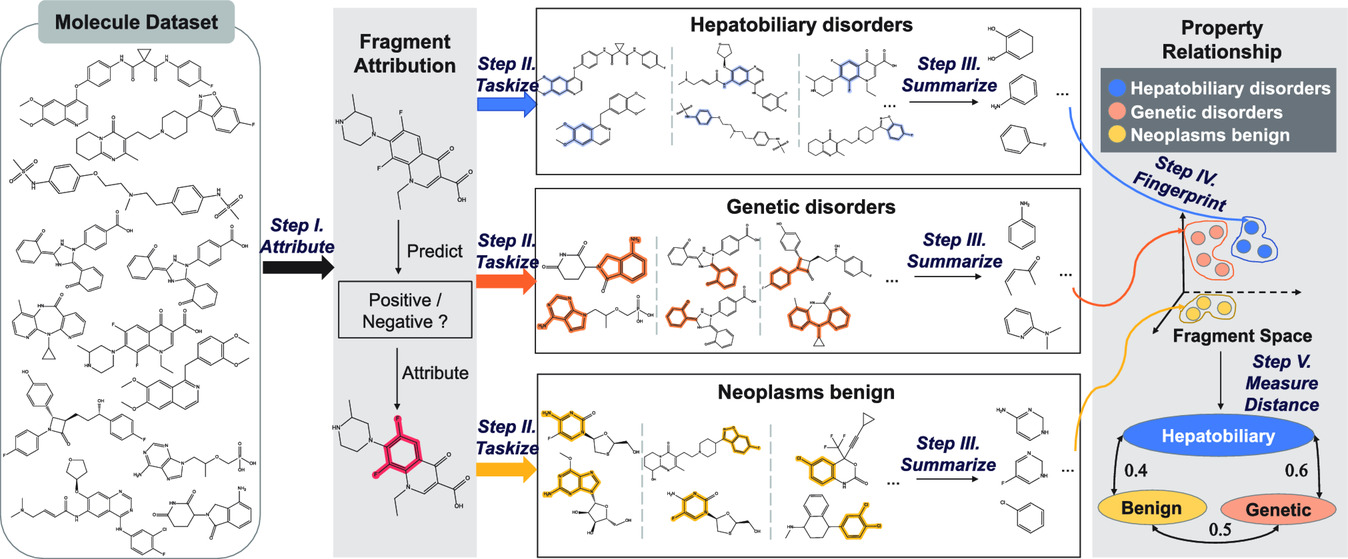
Explainable Fragment-Based Molecular Property Attribution
Linxiang Jia, Zunlei Feng, Odin Zhang, Jie Song, Zipeng Zhong, Shaolun Yao, Mingli Song.
Adv. Intell. Syst. 4(10), 2200104
I led the experimental validation of the model throught collection of pharmacological data, this is my early-stage participation in undergraduate period
Classical CADD and Molecular Dynamics
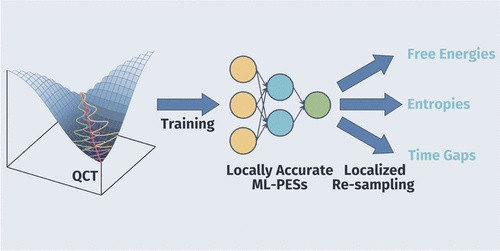
Quasiclassical Trajectory Simulation as a Protocol to Build Locally Accurate Machine Learning Potentials
Jintu Zhang*, Odin Zhang*, Zhixin Qin, Yu Kang, Xin Hong, Tingjun Hou.
J. Chem. Inf. Model. 2023, 63, 4, 1133–1142
We show that quasiclassical trajectory (QCT) calculations can be invoked to efficiently obtain locally accurate ML-PESs, thus accelearing molecular dynamics
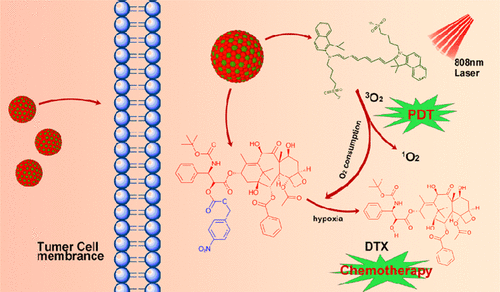
An Engineered Design of Self-Assembly Nanomedicine Guided by Molecular Dynamic Simulation for Photodynamic and Hypoxia-Directed Therapy
Yu Wang, Odin Zhang, Jianwei Wang, Guping Tang, Hongzhen Bai.
Mol. Pharmaceutics, 2023, 20, 4, 2128–2137
I conduct the MD and DPD to guide the design of the nanomedicine ISDNN, which achieves uniform size distribution and high drug loading up to 90%
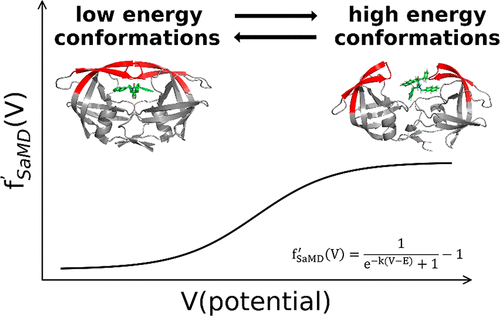
Sigmoid Accelerated Molecular Dynamics: An Efficient Enhanced Sampling Method for Biosystems
Yihao Zhao, Jintu Zhang, Odin Zhang, Shukai Gu, Yafeng Deng, Yaoquan Tu, Tingjun Hou, Yu Kang.
J. Phys. Chem. Lett. 2023, 14, 4, 1103–1112
We propose sigmoid accelerated molecular dynamics (SaMD), which can produce the free energy landscape with better accuracy and efficiency compared with GaMD.
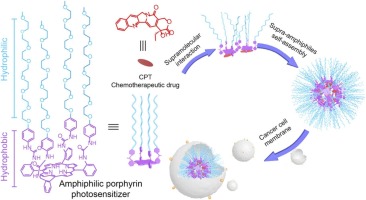
Amphiphilic porphyrin-based supramolecular self-assembly for photochemotherapy: From molecular design to application
Shuping Wang, Xuechun Huang, Yunhong He, Odin Zhang*, Jun Zhou, Guping Tang, Shijun Li, Hongzhen Bai.
Nano Today, 48, 101732
I conduct the Disspative Particle Dynamics (DPD) to study the stability of CPT@P complex, this is my early-stage work in undergraduate period
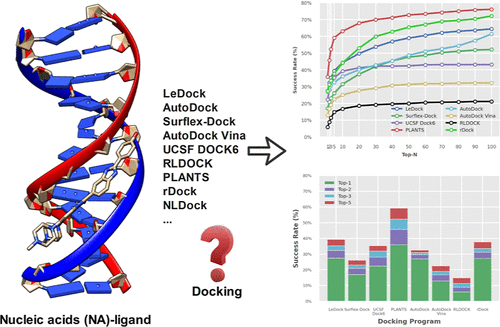
How Good Are Current Docking Programs at Nucleic acids-ligand Docking? a Comprehensive Evaluation
Dejun Jiang, Huifeng Zhao, ...,Odin Zhang, Tingjun Hou
Journal of Chemical Theory and Computation 14(8), 2054-2069
We conducted a comprehensive evaluation of molecular docking programs for NA–ligand systems, contributing to the future research about virtual screening on such a system
Research_Blog
Research_Blog could be found in my Github
RDKit Blog, contains the dihedral finding, conformation generation, even electronic density computation!
PDB Blog, contains the use of PDBParser, you could find the cripts for downloading AlphaFold-Predicted Structures, PDBfile->fasta format, and even sequence alignment!
Matplotlib Blog, a informative figure make it easier for readers to understand research data.
Zan/Pessimistic/Romantic
Dedicate to HanShan and The Dharma Bums

山中何太冷,自古非今年。
沓嶂恒凝雪,幽林每吐烟。
草生芒种后,叶落立秋前。
此有沉迷客,窥窥不见天。
It's cold in the mountains, and not just this year.
The jagged cliffs are always piled high with snow, and the deep canyon species of trees are always spitting mist outward.
The grass is still sprouting in June, and the leaves start falling in August when they first arrive.
And here I was, mesmerized like a drug addict who had just finished a hit.

从明天起,做一个幸福的人
喂马,劈柴,周游世界
从明天起,关心粮食和蔬菜
我有一所房子,面朝大海,春暖花开
从明天起,和每一个亲人通信
告诉他们我的幸福
那幸福的闪电告诉我的
我将告诉每一个人
给每一条河每一座山取一个温暖的名字
陌生人,我也为你祝福
愿你有一个灿烂的前程
愿你有情人终成眷属
愿你在尘世获得幸福
我只愿面朝大海,春暖花开
Face to the sea,spring blossoms
From tomorrow,be a felicific man.
Feed a horse,split firewood,
and travel around the world.
From tomorrow,only care about cereal and vegerable.
I have a house facing to the sea.
There is spring all the year around and all is blooming.
From tomorrow,communciate with every kinsfolk.
Tell them my happiness.
What the felicific lightning has told me is
what I will tell everyone.
Give every river and every mountain a fragant name.
Stranger,I also bless you.
Hope you have boundless prospects.
Hope you have your Jack or Jill.
Hope you find your happiness in the world.
I only shall face to the sea and long for spring blossoms.

A conversation at the bottom of the heart
You have fulfilled your long-standing dream, and broken through all obstacles.
Why do you still feel so painful?
At the moment I realise my dream,
I really felt great satisfaction at the bottom of my heart.
But almost in an instant,
I felt the endless nothingness.
What did the aurora bring to me?
There seems to be nothing.
I still clearly remember the palpitation in my heart when I dreamed of seeing the aurora as a child,
but when I grew up, the throbbing became lighter and lighter.
I was overwhelmed by life, and was no longer able to complete the so-called dream.
But when I buried my dream,
my heart was unwilling.
So, I' ve tried my best,
and came here with the body that I didn't know how long I could live,
I long for the aurora to bring me an eternal meaning,
so that I can fight (against) life bravely.
But that doesn't exist.
It has no meaning at all.
My life will not change, and my story will not fly to the blue sky.
In that case, what is the meaning of life?
The dream that can be moved for it will be dull after it is realized.
All our pursues will turn into nothingness in the end.
Why should we purse it?
So, I'm not going to go on.
Since life has no meaning, let me go.
Let me go to the abyss, to embrace the eternal nothingness,
to embrace the eternal death.
